
( Note: Cite this publication for CellProfiler for cells. CellProfiler 3.0: Next-generation image processing for biology. For single-cell tracking, we used the TrackObjects module in CellProfiler on the.

( Note: Cite this publication for CellProfiler for cells.)Ĭ. 3A, see Supplemental Text for details) and tracked individual cell. Mercurial > repos > bgruening > cpcellprofiler view trackobjects.py 6: 0e4dccaafef5 draft default tip Find changesets by keywords (author, files, the commit message), revision number or hash, or revset expression.
Cellprofiler tracking software#
CellProfiler: image analysis software for identifying and quantifying cell phenotypes. It covers various aspects of CellStar algorithm usage within CellProfiler.

Time-lapse assays probe biological questions that can only be investigated. Since the segmentation and tracking of cells in brightfield images is. Time-lapse analysis of cellular images is an important and growing need in biology. Many open source solutions such as Icy and CellProfiler exist. CellProfiler Tracer: exploring and validating high-throughput, time-lapse microscopy image data Abstract. and CellProfiler for (1) studying cell morphology and migration via tracking. Recent progress on deep learning dramatically improved cell segmentation and tracking. ImageJ and CellProfiler: Complements in Open-Source Bioimage Analysis.
Cellprofiler tracking install#
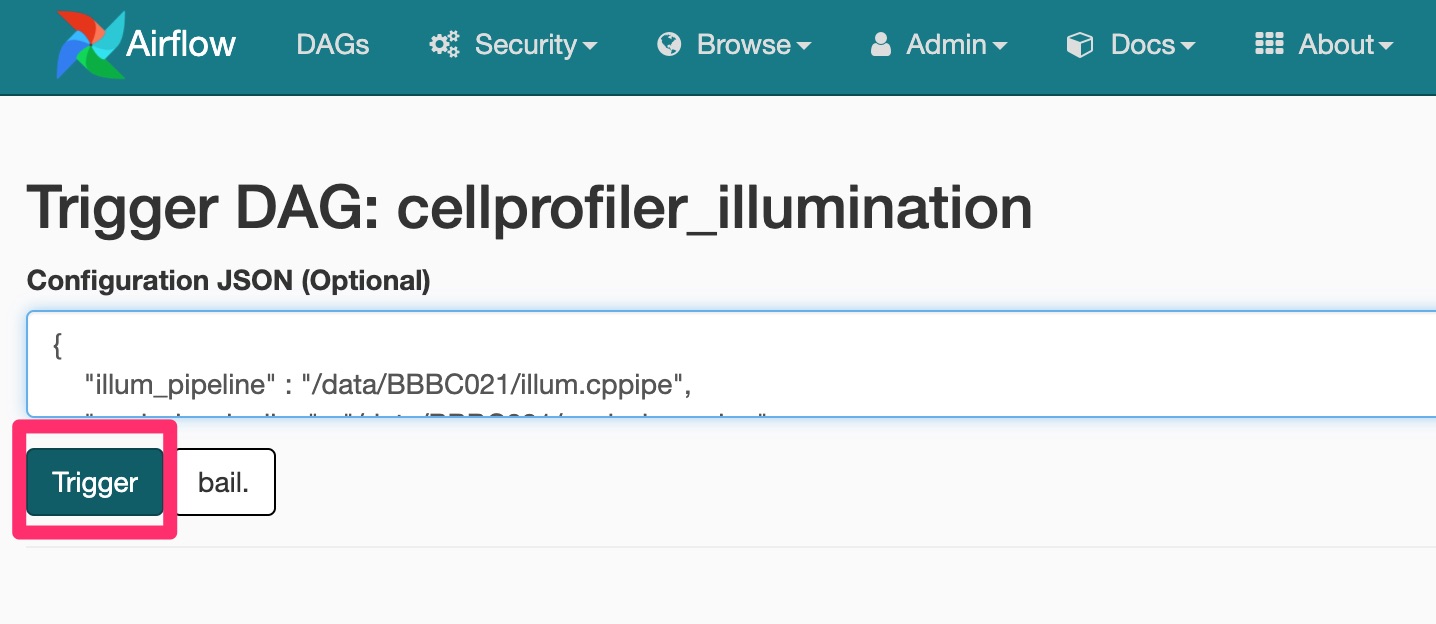
Is a cell image analysis software designed to enable biologists without training in computer vision or programming to quantitatively measure phenotypes from thousands of images automatically.


 0 kommentar(er)
0 kommentar(er)
